I do all my academic writing in R Markdown. This makes my work more transparent and reproducible for others and my future self.
Along the way, I have often needed to use a specific LaTeX template from some conference to format my PDF output from R Markdown. Over time, I’ve learned that there are 4 steps involved in doing this.
In this post, I go over these four steps, and illustrate them with the PLOS LaTeX template.
Step 1: Make the template compile on its own (aka, watch your $)
The first step is to do the simplest thing you can. Download the LaTeX template files from your journal/conference. Then create a new R Markdown document, tell it to use the .tex file from PLOS as template for PDF output, and try to knit.
For example, if you go and download the PLOS LaTeX template, we get a folder that includes the file plos_latex_template.tex. Let’s create a new R Markdown file with this content, and knit to PDF:
---
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
---Argh, we get an error message:
Error compiling template "plos_latex_template.tex" (line 57, column 121):
unexpected "2"
expecting "$"
Error: pandoc document conversion failed with error 5
Execution haltedWhat’s going on? It turns out that dollar signs ($) have special meaning for pandoc, the program responsible for part of the journey from R Markdown to PDF. When pandoc sees a dollar sign in your template, it thinks you want to plug in content from your R Markdown file.
How do we solve this? Throw more money at the problem! That is, for pandoc, a dollar sign in your template is escaped with another dollar sign. So let’s replace all single $ with $$ in plos_latex_template.tex.
If you now knit to PDF, you get the expected output:
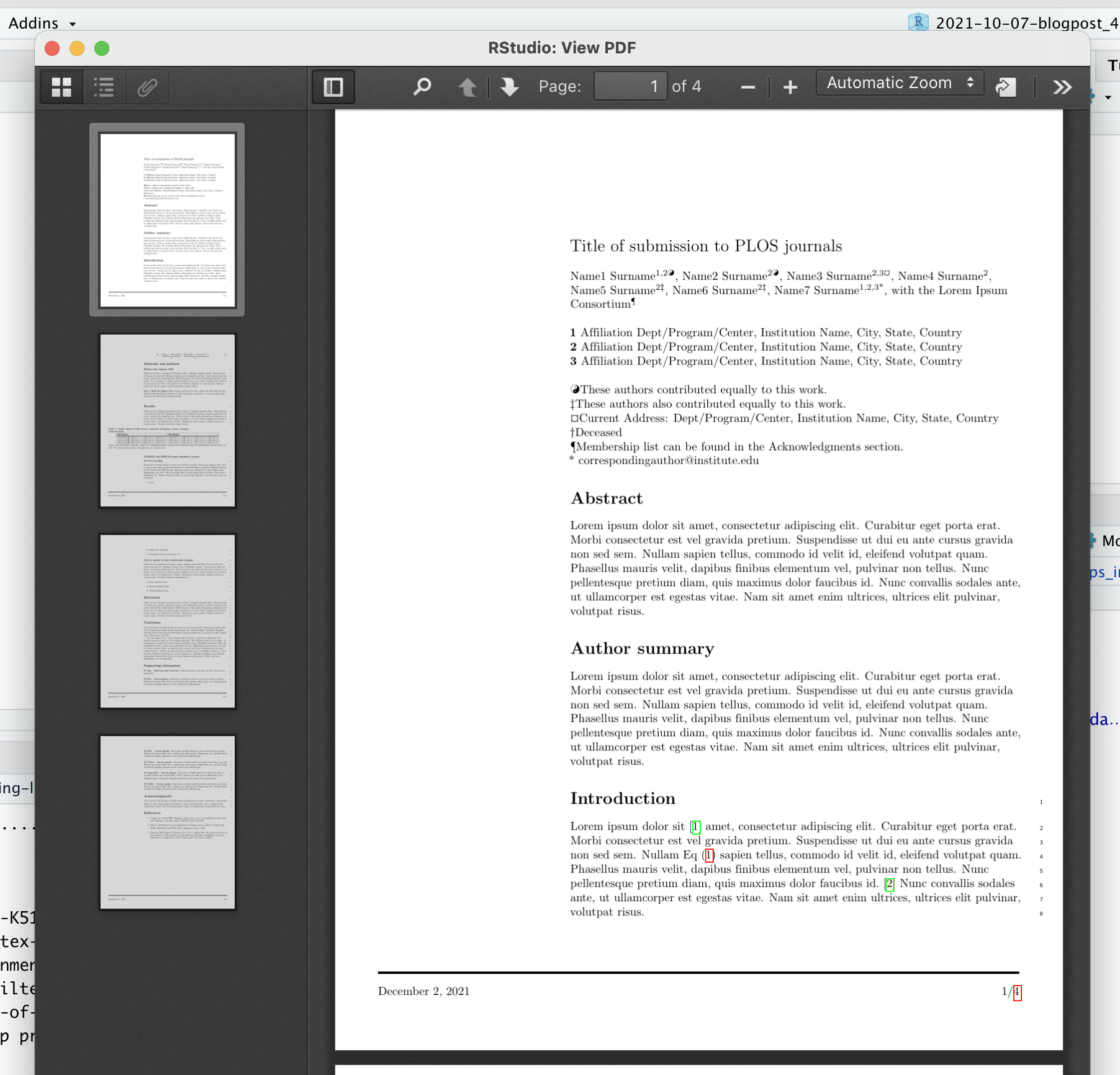
Awesome! Our R Markdown file compiled using PLOS’ .tex file as a template!
Step 2: Pull in desired content from the R Markdown file
The only content we see in the PDF is the sample content from PLOS’ .tex template. How do we insert content from our R Markdown file? Use dollar signs.
When pandoc sees something in the .tex file surrounded by single dollar signs, it’ll look for something that corresponds to it in the R Markdown file. Ask yourself this:
- What and where do you want to plug in stuff from your YAML header (i.e., the stuff in between
---at the top of your R Markdown file)? - Where do you want to plug in the body content?
Pulling in stuff from the YAML header
We can start by simply adding a title to our R Markdown file:
---
title: "R Markdown + LaTeX Templates = <3"
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
---Then open up plos_latex_template.tex and look for the title. It’s here:
% Title must be 250 characters or less.
\begin{flushleft}
{\Large
\textbf\newline{Title of submission to PLOS journals} % Please use "sentence case" for title and headings (capitalize only the first word in a title (or heading), the first word in a subtitle (or subheading), and any proper nouns).
}
\newlineChange it to this:
% Title must be 250 characters or less.
\begin{flushleft}
{\Large
\textbf\newline{$title$} % Please use "sentence case" for title and headings (capitalize only the first word in a title (or heading), the first word in a subtitle (or subheading), and any proper nouns).
}
\newlineNow, if we knit we get this:

The variable names in our YAML header are arbitrary - they don’t have to be something that pandoc has seen before.
Let’s pull in also the abstract and the author summary, from the R Markdown file, and use silly variable names we make up:
---
title: "R Markdown + LaTeX Templates = <3"
silly-abstract: "This is the greatest and best abstract in the world."
silly-author-summary: "The important thing to know about this paper is that it is was written in close collaboration with Tenacious D, without whom our academic universe would have ceased to be."
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
---Find the abstract and author summary sections in plos_latex_template.tex and replace the sample content with our silly variable names:
% Please keep the abstract below 300 words
\section*{Abstract}
$silly-abstract$
% Please keep the Author Summary between 150 and 200 words
% Use first person. PLOS ONE authors please skip this step.
% Author Summary not valid for PLOS ONE submissions.
\section*{Author summary}
$silly-author-summary$When we knit we get the expected output:

Pulling in the body content
Let’s add a bit of body content to our R Markdown file:
---
title: "R Markdown + LaTeX Templates = <3"
silly-abstract: "This is the greatest and best abstract in the world."
silly-author-summary: "The important thing to know about this paper is that it is was written in close collaboration with Tenacious D, without whom our academic universe would have ceased to be."
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
---
# Hello, LaTeX template!
This is a test.The body content is pulled in with a special variable: wherever you write $body$ in the template, pandoc will insert the body content from our R Markdown file.
In our example with the PLOS template, try replacing everything from the introduction to end of the document with the body content. So
% Use "Eq" instead of "Equation" for equation citations.
\section*{Introduction}
Lorem ipsum dolor sit~\cite{bib1} amet, consectetur adipiscing elit
etc...becomes just
$body$When we knit we get:

Step 3: Enable code inclusion
Notice that if we add a code chunk in our R Markdown file, with the chunk option echo=TRUE, we get an error:
---
title: "R Markdown + LaTeX Templates = <3"
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
---
# Hello, LaTeX template!
This is a test.
```{r, echo=TRUE}
2 + 2
```Here’s the error:
! LaTeX Error: Environment Shaded undefined.
Error: LaTeX failed to compile using-latex-templates.tex. See https://yihui.org/tinytex/r/#debugging for debugging tips. See using-latex-templates.log for more info.
Execution haltedAs the error message suggests, it’s because code chunks in your R markdown file gets included in the LaTeX file in a ‘Shaded’ environment (our PDF output is generated from the intermediary LaTeX file).
If you add keep_tex: true to your R Markdown file’s YAML header and knit again, then it’ll keep the generated LaTeX file, so you can have a look at what went wrong:
---
title: "R Markdown + LaTeX Templates = <3"
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
keep_tex: true
---If you find the place in the LaTeX file where the code chunk would appear, it looks like this:
\begin{Shaded}
\begin{Highlighting}[]
\DecValTok{2} \SpecialCharTok{+} \DecValTok{2}
\end{Highlighting}
\end{Shaded}The problem is that our template .tex file doesn’t define this Shaded environment.
We can fix it by adding these three lines to the LaTeX template, somewhere before the \begin{document} command:
$if(highlighting-macros)$
$highlighting-macros$
$endif$What this will do is that if our R Markdown document includes code chunks with echo=TRUE (so that the code is displayed), then the necessary LaTeX packages and environments are automatically added.
If we knit again, we see the expected output:

Step 4: Decide how to include citations
Finally, you need to decide how you want to include citations. There are (at least) three options:
- Use CSL (Citation Style Language) references
- Use the
natbibLaTeX package - Use the
biblatexLaTeX package
Let’s look at each.
CSL references
The Citation Style Language is an open-source project supported by Zotero, Mendeley, RefWorks, and others, and is the approach pandoc uses by default.
To enable it, you need to add the following lines to your LaTeX template:
$if(csl-refs)$
\newlength{\cslhangindent}
\setlength{\cslhangindent}{1.5em}
\newlength{\csllabelwidth}
\setlength{\csllabelwidth}{3em}
\newlength{\cslentryspacingunit} % times entry-spacing
\setlength{\cslentryspacingunit}{\parskip}
\newenvironment{CSLReferences}[2] % #1 hanging-ident, #2 entry spacing
{% don't indent paragraphs
\setlength{\parindent}{0pt}
% turn on hanging indent if param 1 is 1
\ifodd #1
\let\oldpar\par
\def\par{\hangindent=\cslhangindent\oldpar}
\fi
% set entry spacing
\setlength{\parskip}{#2\cslentryspacingunit}
}%
{}
\usepackage{calc}
\newcommand{\CSLBlock}[1]{#1\hfill\break}
\newcommand{\CSLLeftMargin}[1]{\parbox[t]{\csllabelwidth}{#1}}
\newcommand{\CSLRightInline}[1]{\parbox[t]{\linewidth - \csllabelwidth}{#1}\break}
\newcommand{\CSLIndent}[1]{\hspace{\cslhangindent}#1}
$endif$(These commands are taken from the default pandoc LaTeX template and is how pandoc usually tells LaTeX how to include references).
Let’s try it! Add the lines to the PLOS .tex template (somewhere before the \begin{document} command), then create a new file called references.bib and save it with this entry, for illustration purposes:
@InProceedings{lyngs2019,
title = {Self-{{Control}} in {{Cyberspace}}: Applying {{Dual Systems Theory}} to a {{Review}} of {{Digital Self}}-{{Control Tools}}},
booktitle = {{{CHIConference}} on {{Human Factors}} in {{Computing Systems Proceedings}} ({{CHI}} 2019)},
author = {Ulrik Lyngs and Kai Lukoff and Petr Slovak and Reuben Binns and Adam Slack and Michael Inzlicht and Max {Van Kleek} and Nigel Shadbolt},
date = {2019},
publisher = {{ACM}},
location = {{New York, NY, USA}},
doi = {10.1145/3290605.3300361}
}Now point to this file as your bibliography in your R Markdown file’s YAML header, add a citation to lyngs2019 in the body text, and add a ‘References’ heading by the end of the document:
---
title: "R Markdown + LaTeX Templates = <3"
silly-abstract: "This is the greatest and best abstract in the world."
silly-author-summary: "The important thing to know about this paper is that it is was written in close collaboration with Tenacious D, without whom our academic universe would have ceased to be."
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
bibliography: references.bib
---
# Hello, LaTeX template!
This is a test.
This is a citation: @lyngs2019
```{r, echo=TRUE}
2 + 2
```
# ReferencesWhen we knit, we get the expected output:

If we inspect the generated .tex file, we can see that the citation text appears as plain text in the body content:
This is a citation: Lyngs et al. (2019)And the references section were generated by embedding the citation in a CSLReferences environment:
\begin{CSLReferences}{1}{0}
\leavevmode\vadjust pre{\hypertarget{ref-lyngs2019}{}}%
Lyngs, Ulrik, Kai Lukoff, Petr Slovak, Reuben Binns, Adam Slack, Michael Inzlicht, Max Van Kleek, and Nigel Shadbolt. 2019. {``Self-{Control} in {Cyberspace}: Applying {Dual Systems Theory} to a {Review} of {Digital Self}-{Control Tools}.''} In \emph{{CHIConference} on {Human Factors} in {Computing Systems Proceedings} ({CHI} 2019)}. {New York, NY, USA}: {ACM}. \url{https://doi.org/10.1145/3290605.3300361}.
\end{CSLReferences}To change the citation style, download a csl file from the Zotero Style Repository that corresponds to what you want, and add reference it in your R Markdown file’s YAML header with csl: your-csl-file.csl.
Natbib
To use the natbib package for referencing, first add citation_package: natbib in the YAML header of your R Markdown file:
...
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
citation_package: natbib
bibliography: references.bibThen, in the LaTeX template:
- add
\usepackage[options]{natbib}, and set a citation style with\bibliographystyle{stylename}, somewhere before\begin{document}. - point to your bibliography with
\bibliography{your-reference-file}before\end{document}.
In our example, the PLOS template comes with a style sheet in their plos2015.bst file, so we’ll use that.
\usepackage[numbers]{natbib}
\bibliographystyle{plos2015.bst}
...etc.
\bibliography{$bibliography$}
\end{document}The PLOS template also includes the cite package. Comment this out, because we’re now using natbib instead.
% cite package, to clean up citations in the main text. Do not remove.
% \usepackage{cite}
When we knit, we get this:

If you inspect the .tex file generating the PDF, we can see that the citation is generated with the \citep command, and that the bibliography is simply generated with a \bibliography call.
This is a citation: \citep{lyngs2019}
...
\bibliography{references.bib}Biblatex
To use biblatex for citations, first add citation_package: biblatex in the YAML header of your R Markdown file:
...
output:
bookdown::pdf_document2:
template: plos_latex_template.tex
citation_package: biblatex
bibliography: references.bibThen, in the LaTeX template:
- add
\usepackage{biblatex}and point to your bibliography with\addbibresource{your-references-file}, somewhere before\begin{document}. - add the command
\printbibliographywhere you want the references section to be printed.
\usepackage{biblatex}
\addbibresource{$bibliography$}
...
\printbibliography
\end{document}In the specific case of the PLOS template, it turns out we also need to change \usepackage[utf8x]{inputenc} to \usepackage[utf8]{inputenc}, for reasons explained on StackOverflow.
When we knit, we get this:

If we inspect the .tex file generating the PDF, we can see that our citation is inserted with the \autocite command, and the bibliography is simply generated with the \printbibliography command.
This is a citation: \autocite{lyngs2019}
...
\printbibliographyThat’s it! Go and change the LaTeX world with R Markdown!
Comments
I use the open source, zero-tracking utteranc.es widget for comments - it's built on GitHub issues, so you need a GitHub account to comment.